Bank Customer Satisfaction Prediction Using CNN and Feature Selection
Feature Selection and CNN
In this project we are going to build a neural network to predict if a particular bank customer is satisfies or not. To do this we are going to use Convolutional Neural Networks. The dataset which we are going to use contains 370 features. We are going to use feature selection to select the most relevant features and reduce the complexity of our model.
Dataset
The dataset is an anonymized dataset which contains a large number of numeric variables. The TARGET column is the variable to predict. It equals one for unsatisfied customers and 0 for satisfied customers.
You can download the dataset from any one of the links given below:-
https://www.kaggle.com/c/santander-customer-satisfaction/data
https://github.com/laxmimerit/Data-Files-for-Feature-Selection
We are going to use tensorflow to build the LSTM. You can install tensorflow by running this command. If you machine has a GPU you can use the second command.
!pip install tensorflow
!pip install tensorflow-gpu
The necessary python libraries are imported here-
numpyis used to perform basic array operationsseabornis used to visualize the resultspandasfor loading and manipulating the data.pyplotfrom matplotlib is used to visualize the results.train_test_splitis used to split the data into training and testing datasets.StandardScaleris used to scale the values in the data.VarianceThresholdis used for feature selection.Tensorflowis used to build the neural network.- We have even imported all the layers required to build the model from
keras.
import numpy as np import pandas as pd import seaborn as sns import matplotlib.pyplot as plt from sklearn.model_selection import train_test_split from sklearn.preprocessing import StandardScaler from sklearn.feature_selection import VarianceThreshold import tensorflow as tf from tensorflow.keras import Sequential from tensorflow.keras.layers import Conv1D, MaxPool1D, Flatten, Dense, Dropout, BatchNormalization from tensorflow.keras.optimizers import Adam print(tf.__version__)
2.1.0
You can use this command to directly get the data from github.
!git clone https://github.com/laxmimerit/Data-Files-for-Feature-Selection.git
After downloading the data, we will now read the data using read_csv(). To see the first 5 rows of the data we can use data.head().
data = pd.read_csv('train.csv')
data.head()
| ID | var3 | var15 | imp_ent_var16_ult1 | imp_op_var39_comer_ult1 | imp_op_var39_comer_ult3 | imp_op_var40_comer_ult1 | imp_op_var40_comer_ult3 | imp_op_var40_efect_ult1 | imp_op_var40_efect_ult3 | ... | saldo_medio_var33_hace2 | saldo_medio_var33_hace3 | saldo_medio_var33_ult1 | saldo_medio_var33_ult3 | saldo_medio_var44_hace2 | saldo_medio_var44_hace3 | saldo_medio_var44_ult1 | saldo_medio_var44_ult3 | var38 | TARGET | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 23 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 39205.170000 | 0 |
| 1 | 3 | 2 | 34 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 49278.030000 | 0 |
| 2 | 4 | 2 | 23 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 67333.770000 | 0 |
| 3 | 8 | 2 | 37 | 0.0 | 195.0 | 195.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 64007.970000 | 0 |
| 4 | 10 | 2 | 39 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 117310.979016 | 0 |
5 rows × 371 columns
We have 76020 rows in the dataset and 371 columns.
data.shape
(76020, 371)
Now we are going to create a feature space X. Feature space will only contain the column which provide information necessary for prediction. ID and TARGET do not play any role in prediction, so we are going to remove them using drop(). After droppring the 2 columns you can see that the number of columns have reduced to 369.
X = data.drop(labels=['ID', 'TARGET'], axis = 1) X.shape
(76020, 369)
Lets create a variable y which contains the values which have to be predicted i.e. TARGET.
y = data['TARGET']
Now we will split the data into training and testing set with the help of train_test_split(). test_size = 0.2 will keep 20% data for testing and 80% data will be used for training the model. random_state controls the shuffling applied to the data before applying the split. stratify = y means that the data is split in a stratified fashion, using y as the class labels.
X_train, X_test, y_train, y_test = train_test_split(X,y, test_size = 0.2, random_state = 0, stratify = y)
As we can see, the training dataset consists of 60816 rows i.e. 80% of the data and the testing dataset consists of 15204 rows i.e 20% of the data.
X_train.shape, X_test.shape
((60816, 369), (15204, 369))
Remove Constant, Quasi Constant and Duplicate Features
Feature selection is the process of reducing the number of input variables when developing a predictive model.
Constant Featuresare the features that show single values in all the observations in the dataset. These features provide no information that allows ML models to predict the target.Quasi constantfeatures, as the name suggests, are the features that are almost constant. In other words, these features have the same values for a very large subset of the outputs. They have less variance. Such features are not very useful for making predictions.Duplicate Featuresas the name suggests are duplicated in the dataset.
Here we have set the variance threshold to 1% i.e. if any column has variance less than 1% it will be removed. In other words only the columns having variance greater than 99% will be retained. We are fitting VarianceThreshold() to the training data and not the test data. We are only transforming the test data.
filter = VarianceThreshold(0.01) X_train = filter.fit_transform(X_train) X_test = filter.transform(X_test) X_train.shape, X_test.shape
((60816, 273), (15204, 273))
After removing the Quasi constant features we can see that 96 features are removed from the dataset.
369-273
96
Now we will remove the duplicate features. We don't have any direct function to remove duplicate features but we have functions to check for duplicate rows. Hence we are taking transpose of the data set using .T. As we can see after taking transpose the shape of X_train_T is exactly opposite to that of X_train.
X_train_T = X_train.T X_test_T = X_test.T X_train_T = pd.DataFrame(X_train_T) X_test_T = pd.DataFrame(X_test_T) X_train_T.shape
(273, 60816)
.duplicated() returns a boolean Series denoting duplicate rows. We can see that 17 features are duplicated.
X_train_T.duplicated().sum()
17
Now we will see the list of dupicated features. The features having index True are duplicated.
duplicated_features = X_train_T.duplicated() duplicated_features[70:90]
70 False 71 False 72 True 73 False 74 True 75 False 76 False 77 False 78 False 79 False 80 False 81 False 82 False 83 False 84 False 85 False 86 False 87 False 88 False 89 False dtype: bool
We have to retain the features with False value because they are not duplicated. So here we are going to use inversion i.e. we are going to change the False value to True and viceversa.
features_to_keep = [not index for index in duplicated_features] features_to_keep[70:90]
[True, True, False, True, False, True, True, True, True, True, True, True, True, True, True, True, True, True, True, True]
Now as we have inverted the values, we have to retain the features with value True. We also have to take transpose once again to get the data back in original shape. Here we have done it for X_train.
X_train = X_train_T[features_to_keep].T X_train.shape
(60816, 256)
Here we have done it for X_test.
X_test = X_test_T[features_to_keep].T X_test.shape
(15204, 256)
X_train.head()
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 263 | 264 | 265 | 266 | 267 | 268 | 269 | 270 | 271 | 272 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2.0 | 26.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 117310.979016 |
| 1 | 2.0 | 23.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 85472.340000 |
| 2 | 2.0 | 23.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 317769.240000 |
| 3 | 2.0 | 30.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 76209.960000 |
| 4 | 2.0 | 23.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | ... | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 302754.000000 |
5 rows × 256 columns
Now we are going to get the bring the data into the same range. StandardScaler() standardizes the features by removing the mean and scaling to unit variance.
scaler = StandardScaler() X_train = scaler.fit_transform(X_train) X_test = scaler.transform(X_test) X_train
array([[ 3.80478472e-02, -5.56029626e-01, -5.27331414e-02, ...,
-1.87046327e-02, -1.97720391e-02, 3.12133758e-03],
[ 3.80478472e-02, -7.87181903e-01, -5.27331414e-02, ...,
-1.87046327e-02, -1.97720391e-02, -1.83006062e-01],
[ 3.80478472e-02, -7.87181903e-01, -5.27331414e-02, ...,
-1.87046327e-02, -1.97720391e-02, 1.17499225e+00],
...,
[ 3.80478472e-02, 5.99731758e-01, -5.27331414e-02, ...,
-1.87046327e-02, -1.97720391e-02, -2.41865113e-01],
[ 3.80478472e-02, -1.70775831e-01, -5.27331414e-02, ...,
-1.87046327e-02, -1.97720391e-02, 3.12133758e-03],
[ 3.80478472e-02, 2.91528722e-01, 7.65192053e+00, ...,
-1.87046327e-02, -1.97720391e-02, 3.12133758e-03]])
X_train.shape, X_test.shape
((60816, 256), (15204, 256))
Our data is 2 dimensional but neural networks accept 3 dimensional data. So we have to reshape() the data.
X_train = X_train.reshape(60816, 256,1) X_test = X_test.reshape(15204, 256, 1) X_train.shape, X_test.shape
((60816, 256, 1), (15204, 256, 1))
y_train = y_train.to_numpy() y_test = y_test.to_numpy()
Building the CNN
A Sequential() model is appropriate for a plain stack of layers where each layer has exactly one input tensor and one output tensor.
Conv1D() is a 1D Convolution Layer, this layer is very effective for deriving features from a fixed-length segment of the overall dataset, where it is not so important where the feature is located in the segment. In the first Conv1D() layer, we are learning a total of 36 filters with size of the convolutional window as 3. The input_shape specifies the shape of the input. It is a necessary parameter for the first layer in any neural network. We will be using the ReLu activation function. The rectified linear activation function or ReLU for short is a piecewise linear function that will output the input directly if it is positive, otherwise, it will output zero.
BatchNormalization() allows each layer of a network to learn by itself a little bit more independently of other layers. To increase the stability of a neural network, batch normalization normalizes the output of a previous activation layer by subtracting the batch mean and dividing by the batch standard deviation. It applies a transformation that maintains the mean output close to 0 and the output standard deviation close to 1.
MaxPool1D() downsamples the input representation by taking the maximum value over the window defined by pool_size which is 2 in case of the first Max Pool layer of this neural network.
Dropout() is used to randomly set the outgoing edges of hidden units to 0 at each update of the training phase. The value passed in dropout specifies the probability at which outputs of the layer are dropped out.
Flatten() is used to convert the data into a 1-dimensional array for inputting it to the next layer.
Dense() is the regular deeply connected neural network layer. The output layer is a dense layer with 1 neuron because we are predicting a single value. Sigmoid function is used because it exists between (0 to 1) and this facilitates us to predict a binary input.
model = Sequential() model.add(Conv1D(32, 3, activation='relu', input_shape = (256,1))) model.add(BatchNormalization()) model.add(MaxPool1D(2)) model.add(Dropout(0.3)) model.add(Conv1D(64, 3, activation='relu')) model.add(BatchNormalization()) model.add(MaxPool1D(2)) model.add(Dropout(0.5)) model.add(Conv1D(128, 3, activation='relu')) model.add(BatchNormalization()) model.add(MaxPool1D(2)) model.add(Dropout(0.5)) model.add(Flatten()) model.add(Dense(256, activation='relu')) model.add(Dropout(0.5)) model.add(Dense(1, activation='sigmoid'))
model.summary()
Model: "sequential" _________________________________________________________________ Layer (type) Output Shape Param # ================================================================= conv1d (Conv1D) (None, 254, 32) 128 _________________________________________________________________ batch_normalization (BatchNo (None, 254, 32) 128 _________________________________________________________________ max_pooling1d (MaxPooling1D) (None, 127, 32) 0 _________________________________________________________________ dropout (Dropout) (None, 127, 32) 0 _________________________________________________________________ conv1d_1 (Conv1D) (None, 125, 64) 6208 _________________________________________________________________ batch_normalization_1 (Batch (None, 125, 64) 256 _________________________________________________________________ max_pooling1d_1 (MaxPooling1 (None, 62, 64) 0 _________________________________________________________________ dropout_1 (Dropout) (None, 62, 64) 0 _________________________________________________________________ conv1d_2 (Conv1D) (None, 60, 128) 24704 _________________________________________________________________ batch_normalization_2 (Batch (None, 60, 128) 512 _________________________________________________________________ max_pooling1d_2 (MaxPooling1 (None, 30, 128) 0 _________________________________________________________________ dropout_2 (Dropout) (None, 30, 128) 0 _________________________________________________________________ flatten (Flatten) (None, 3840) 0 _________________________________________________________________ dense (Dense) (None, 256) 983296 _________________________________________________________________ dropout_3 (Dropout) (None, 256) 0 _________________________________________________________________ dense_1 (Dense) (None, 1) 257 ================================================================= Total params: 1,015,489 Trainable params: 1,015,041 Non-trainable params: 448 _________________________________________________________________
Now we will compile and fit the model. We are using Adam optimizer with 0.00005 learning rate. We will use 10 epochs to train the model. An epoch is an iteration over the entire data provided. validation_data is the data on which to evaluate the loss and any model metrics at the end of each epoch. The model will not be trained on this data. As metrics = ['accuracy'] the model will be evaluated based on the accuracy.
model.compile(optimizer=Adam(lr=0.00005), loss='binary_crossentropy', metrics=['accuracy']) history = model.fit(X_train, y_train, epochs=10, validation_data=(X_test, y_test), verbose=1)
Train on 60816 samples, validate on 15204 samples Epoch 5/10 60816/60816 [==============================] - 111s 2ms/sample - loss: 0.1630 - accuracy: 0.9604 - val_loss: 0.1641 - val_accuracy: 0.9605 Epoch 6/10 60816/60816 [==============================] - 111s 2ms/sample - loss: 0.1599 - accuracy: 0.9603 - val_loss: 0.1595 - val_accuracy: 0.9605 Epoch 7/10 60816/60816 [==============================] - 111s 2ms/sample - loss: 0.1576 - accuracy: 0.9604 - val_loss: 0.1590 - val_accuracy: 0.9604 Epoch 8/10 60816/60816 [==============================] - 111s 2ms/sample - loss: 0.1556 - accuracy: 0.9604 - val_loss: 0.1610 - val_accuracy: 0.9605 Epoch 9/10 60816/60816 [==============================] - 111s 2ms/sample - loss: 0.1536 - accuracy: 0.9604 - val_loss: 0.1558 - val_accuracy: 0.9603 Epoch 10/10 60816/60816 [==============================] - 111s 2ms/sample - loss: 0.1542 - accuracy: 0.9604 - val_loss: 0.1602 - val_accuracy: 0.9599
history gives us the summary of all the accuracies and losses calculated after each epoch.
history.history
{'accuracy': [0.95417327,
0.9592706,
0.95992833,
0.96033937,
0.96037227,
0.9603065,
0.9604052,
0.960438,
0.9603887,
0.9604052],
'loss': [0.21693714527215763,
0.17656464240582592,
0.16882949567384484,
0.16588703954582057,
0.16303560407957227,
0.15994301885150822,
0.15763013028843298,
0.15563193596928912,
0.1535658989747522,
0.1542411554370529],
'val_accuracy': [0.9600763,
0.9600763,
0.96033937,
0.9604052,
0.9604709,
0.9604709,
0.9604052,
0.9604709,
0.9602736,
0.959879],
'val_loss': [0.17092196812710614,
0.1765108920851371,
0.16735200087523436,
0.1662461552617033,
0.16413307644895303,
0.1594827836499469,
0.15897791552088097,
0.16101698756464938,
0.15578439738331923,
0.16016060526129197]}
We will now plot the model accuracy and model loss. In model accuracy we will plot the training accuracy and validation accuracy and in model loss we will plot the training loss and validation loss.
def plot_learningCurve(history, epoch):
# Plot training & validation accuracy values
epoch_range = range(1, epoch+1)
plt.plot(epoch_range, history.history['accuracy'])
plt.plot(epoch_range, history.history['val_accuracy'])
plt.title('Model accuracy')
plt.ylabel('Accuracy')
plt.xlabel('Epoch')
plt.legend(['Train', 'Val'], loc='upper left')
plt.show()
# Plot training & validation loss values
plt.plot(epoch_range, history.history['loss'])
plt.plot(epoch_range, history.history['val_loss'])
plt.title('Model loss')
plt.ylabel('Loss')
plt.xlabel('Epoch')
plt.legend(['Train', 'Val'], loc='upper left')
plt.show()
plot_learningCurve(history, 10)
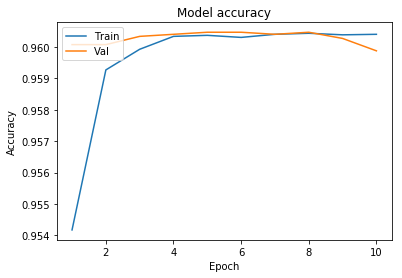
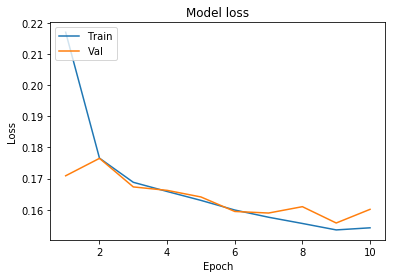
We have got an accuracy of 96%. Hence we can conclude that Convolutional neural networks with appropriate feature selection can build a very powerful model for this dataset. Feature selection enables the machine learning algorithm to train faster. It reduces the complexity of a model and makes it easier to interpret. It also improves the accuracy of a model if the right subset is chosen.
0 Comments