Step Forward, Step Backward and Exhaustive Feature Selection | Wrapper Method | KGP Talkie
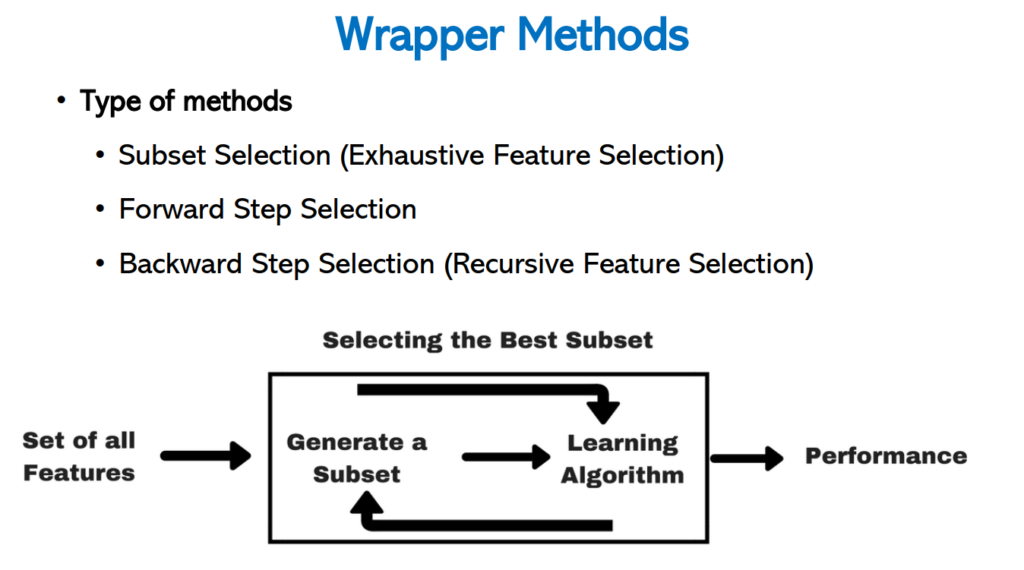
Wrapping method
Uses of Wrapping method
- Use combinations of variables to determine predictive power.
- To find the best combination of variables.
- Computationally expensive than filter method.
- To perform better than filter method.
- Not recommended on high number of features.
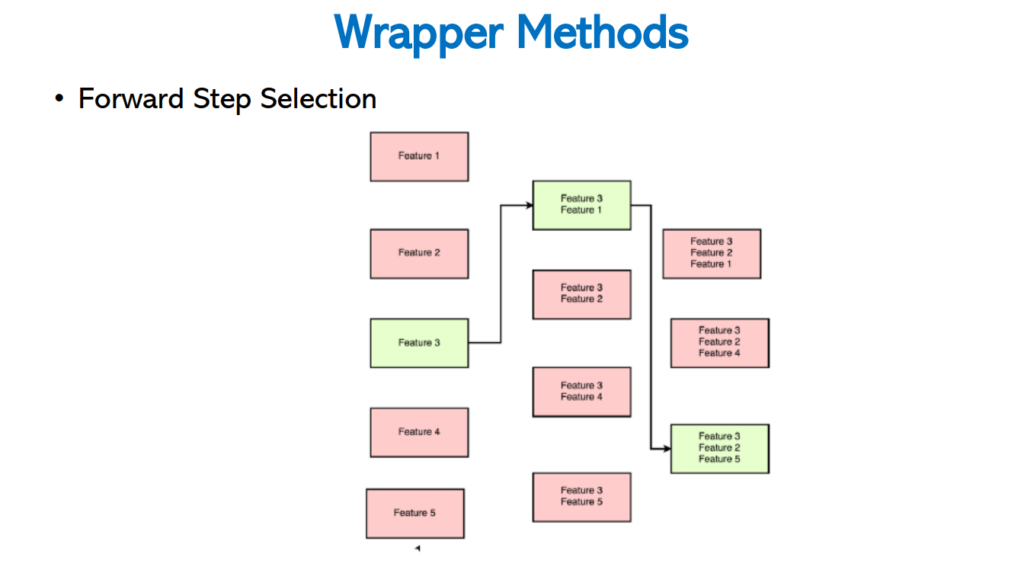
Forward Step Selection
In this wrapping method, it selects one best feature every time and finally it combines all the best features for the best accuracy.
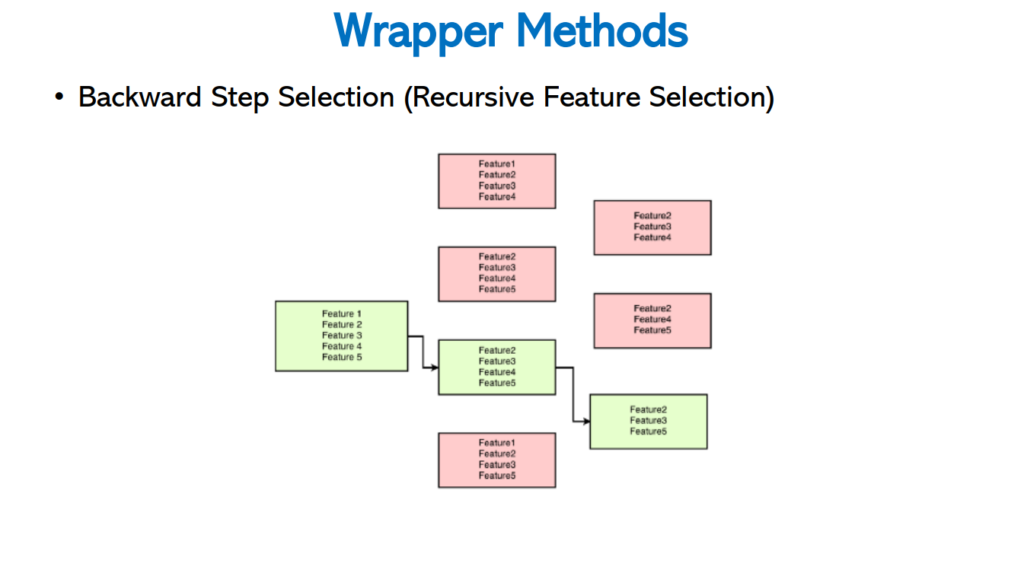
Backward Step Selection
It is reverse process of Forward Step Selection method, intially it takes all the features and remove one by one every time. Finally it left with required number of features for the best accuracy.
Exhaustive Feature Selection
- It is also called as subset selection method.
- It fits the model with each possible combinations of N features.
( y = B0, y = B0 + B1.X1, y = C0 + C1.X2 ) - It requires massive computational power.
- It uses test error to evaluate model performance.
Drawback
It is a slower method compared to step forward and back ward methods.
Use of mlxtend in Wrapper Method
!pip install mlxtend
Requirement already satisfied: mlxtend in c:\users\srish\appdata\roaming\python\python38\site-packages (0.17.3) Requirement already satisfied: scikit-learn>=0.20.3 in e:\callme_conda\lib\site-packages (from mlxtend) (0.23.1) Requirement already satisfied: pandas>=0.24.2 in e:\callme_conda\lib\site-packages (from mlxtend) (1.0.5) Requirement already satisfied: joblib>=0.13.2 in e:\callme_conda\lib\site-packages (from mlxtend) (0.16.0) Requirement already satisfied: matplotlib>=3.0.0 in e:\callme_conda\lib\site-packages (from mlxtend) (3.2.2) Requirement already satisfied: setuptools in e:\callme_conda\lib\site-packages (from mlxtend) (49.2.0.post20200714) Requirement already satisfied: numpy>=1.16.2 in e:\callme_conda\lib\site-packages (from mlxtend) (1.18.5) Requirement already satisfied: scipy>=1.2.1 in e:\callme_conda\lib\site-packages (from mlxtend) (1.5.0) Requirement already satisfied: threadpoolctl>=2.0.0 in e:\callme_conda\lib\site-packages (from scikit-learn>=0.20.3->mlxtend) (2.1.0) Requirement already satisfied: python-dateutil>=2.6.1 in e:\callme_conda\lib\site-packages (from pandas>=0.24.2->mlxtend) (2.8.1) Requirement already satisfied: pytz>=2017.2 in e:\callme_conda\lib\site-packages (from pandas>=0.24.2->mlxtend) (2020.1) Requirement already satisfied: cycler>=0.10 in e:\callme_conda\lib\site-packages (from matplotlib>=3.0.0->mlxtend) (0.10.0) Requirement already satisfied: kiwisolver>=1.0.1 in e:\callme_conda\lib\site-packages (from matplotlib>=3.0.0->mlxtend) (1.2.0) Requirement already satisfied: pyparsing!=2.0.4,!=2.1.2,!=2.1.6,>=2.0.1 in e:\callme_conda\lib\site-packages (from matplotlib>=3.0.0->mlxtend) (2.4.7) Requirement already satisfied: six>=1.5 in e:\callme_conda\lib\site-packages (from python-dateutil>=2.6.1->pandas>=0.24.2->mlxtend) (1.15.0)
More Information Available at http://rasbt.github.io/mlxtend/
How it works
Sequential feature selection algorithms are a family of greedy search algorithms that are used to reduce an initial d-dimensional feature space to a k-dimensional feature subspace where k < d.
In a nutshell, SFAs remove or add one feature at the time based on the classifier performance until a feature subset of the desired size k is reached. There are 4 different flavors of SFAs available via the SequentialFeatureSelector:
- Sequential Forward Selection (SFS)
- Sequential Backward Selection (SBS)
- Sequential Forward Floating Selection (SFFS)
- Sequential Backward Floating Selection (SBFS)
Step Forward Selection (SFS)
Importing required libraries
import numpy as np import pandas as pd import seaborn as sns import matplotlib.pyplot as plt %matplotlib inline
from sklearn.model_selection import train_test_split from sklearn.ensemble import RandomForestRegressor, RandomForestClassifier from sklearn.metrics import roc_auc_score from mlxtend.feature_selection import SequentialFeatureSelector as SFS
from sklearn.datasets import load_wine from sklearn.preprocessing import StandardScaler
We are going to wine dataset. We can load this dataset from sklearn.
data = load_wine()
Let's get the keys of this dataset.
data.keys()
dict_keys(['data', 'target', 'frame', 'target_names', 'DESCR', 'feature_names'])
Let's get the description of the wine dataset.
print(data.DESCR)
.. _wine_dataset:
Wine recognition dataset
------------------------
**Data Set Characteristics:**
:Number of Instances: 178 (50 in each of three classes)
:Number of Attributes: 13 numeric, predictive attributes and the class
:Attribute Information:
- Alcohol
- Malic acid
- Ash
- Alcalinity of ash
- Magnesium
- Total phenols
- Flavanoids
- Nonflavanoid phenols
- Proanthocyanins
- Color intensity
- Hue
- OD280/OD315 of diluted wines
- Proline
- class:
- class_0
- class_1
- class_2
:Summary Statistics:
============================= ==== ===== ======= =====
Min Max Mean SD
============================= ==== ===== ======= =====
Alcohol: 11.0 14.8 13.0 0.8
Malic Acid: 0.74 5.80 2.34 1.12
Ash: 1.36 3.23 2.36 0.27
Alcalinity of Ash: 10.6 30.0 19.5 3.3
Magnesium: 70.0 162.0 99.7 14.3
Total Phenols: 0.98 3.88 2.29 0.63
Flavanoids: 0.34 5.08 2.03 1.00
Nonflavanoid Phenols: 0.13 0.66 0.36 0.12
Proanthocyanins: 0.41 3.58 1.59 0.57
Colour Intensity: 1.3 13.0 5.1 2.3
Hue: 0.48 1.71 0.96 0.23
OD280/OD315 of diluted wines: 1.27 4.00 2.61 0.71
Proline: 278 1680 746 315
============================= ==== ===== ======= =====
:Missing Attribute Values: None
:Class Distribution: class_0 (59), class_1 (71), class_2 (48)
:Creator: R.A. Fisher
:Donor: Michael Marshall (MARSHALL%PLU@io.arc.nasa.gov)
:Date: July, 1988
This is a copy of UCI ML Wine recognition datasets.
https://archive.ics.uci.edu/ml/machine-learning-databases/wine/wine.data
The data is the results of a chemical analysis of wines grown in the same region in Italy by three different cultivators. There are thirteen differentmeasurements taken for different constituents found in the three types of wine.
Let's go ahead and get the data in x and y vectors.
X = pd.DataFrame(data.data) y = data.target
X.columns = data.feature_names X.head()
| alcohol | malic_acid | ash | alcalinity_of_ash | magnesium | total_phenols | flavanoids | nonflavanoid_phenols | proanthocyanins | color_intensity | hue | od280/od315_of_diluted_wines | proline | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 14.23 | 1.71 | 2.43 | 15.6 | 127.0 | 2.80 | 3.06 | 0.28 | 2.29 | 5.64 | 1.04 | 3.92 | 1065.0 |
| 1 | 13.20 | 1.78 | 2.14 | 11.2 | 100.0 | 2.65 | 2.76 | 0.26 | 1.28 | 4.38 | 1.05 | 3.40 | 1050.0 |
| 2 | 13.16 | 2.36 | 2.67 | 18.6 | 101.0 | 2.80 | 3.24 | 0.30 | 2.81 | 5.68 | 1.03 | 3.17 | 1185.0 |
| 3 | 14.37 | 1.95 | 2.50 | 16.8 | 113.0 | 3.85 | 3.49 | 0.24 | 2.18 | 7.80 | 0.86 | 3.45 | 1480.0 |
| 4 | 13.24 | 2.59 | 2.87 | 21.0 | 118.0 | 2.80 | 2.69 | 0.39 | 1.82 | 4.32 | 1.04 | 2.93 | 735.0 |
Now we will chech whether null values present in the dataset by using isnull.sum().
X.isnull().sum()
alcohol 0 malic_acid 0 ash 0 alcalinity_of_ash 0 magnesium 0 total_phenols 0 flavanoids 0 nonflavanoid_phenols 0 proanthocyanins 0 color_intensity 0 hue 0 od280/od315_of_diluted_wines 0 proline 0 dtype: int64
Let's go ahead do the train,test and split for this dataset. Have a look at the following code.
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2, random_state = 0) X_train.shape, X_test.shape
((142, 13), (36, 13))
Let's go ahead and start working for the Step Forward Feature Selection(SFS).
Step Forward Feature Selection (SFS)
Here, we are using SequentialFeatureSelector() and passing Random Forest Classifier in this we are passing number of estimators, random_state and number of jobs.
k number of features are the required number of features.
In this case, since it is forward step method, forward is equal to True.
For verbose it is for log here we are using 2.Cross validation set,here we are choosing as 4.Number of jobs means how many cores we will use, here -1 means use all the available core in this system.
sfs = SFS(RandomForestClassifier(n_estimators=100, random_state=0, n_jobs = -1),
k_features = 7,
forward= True,
floating = False,
verbose= 2,
scoring= 'accuracy',
cv = 4,
n_jobs= -1
).fit(X_train, y_train)
[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 5 out of 13 | elapsed: 4.2s remaining: 6.8s [Parallel(n_jobs=-1)]: Done 13 out of 13 | elapsed: 5.8s finished [2020-08-06 12:56:29] Features: 1/7 -- score: 0.7674603174603174[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 4 out of 12 | elapsed: 1.5s remaining: 3.1s [Parallel(n_jobs=-1)]: Done 12 out of 12 | elapsed: 3.2s finished [2020-08-06 12:56:33] Features: 2/7 -- score: 0.9718253968253968[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 2 out of 11 | elapsed: 2.3s remaining: 10.6s [Parallel(n_jobs=-1)]: Done 8 out of 11 | elapsed: 2.8s remaining: 1.0s [Parallel(n_jobs=-1)]: Done 11 out of 11 | elapsed: 4.7s finished [2020-08-06 12:56:37] Features: 3/7 -- score: 0.9859126984126985[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 7 out of 10 | elapsed: 2.2s remaining: 0.9s [Parallel(n_jobs=-1)]: Done 10 out of 10 | elapsed: 4.5s finished [2020-08-06 12:56:42] Features: 4/7 -- score: 0.9789682539682539[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 4 out of 9 | elapsed: 2.3s remaining: 2.8s [Parallel(n_jobs=-1)]: Done 9 out of 9 | elapsed: 4.1s remaining: 0.0s [Parallel(n_jobs=-1)]: Done 9 out of 9 | elapsed: 4.1s finished [2020-08-06 12:56:46] Features: 5/7 -- score: 0.9720238095238095[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 3 out of 8 | elapsed: 2.1s remaining: 3.6s [Parallel(n_jobs=-1)]: Done 8 out of 8 | elapsed: 2.9s remaining: 0.0s [Parallel(n_jobs=-1)]: Done 8 out of 8 | elapsed: 2.9s finished [2020-08-06 12:56:49] Features: 6/7 -- score: 0.9789682539682539[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 4 out of 7 | elapsed: 2.3s remaining: 1.7s [Parallel(n_jobs=-1)]: Done 7 out of 7 | elapsed: 2.7s finished [2020-08-06 12:56:52] Features: 7/7 -- score: 0.9791666666666666
sfs.k_feature_names_
('alcohol',
'ash',
'magnesium',
'flavanoids',
'proanthocyanins',
'color_intensity',
'proline')
sfs.k_feature_idx_
(0, 2, 4, 6, 8, 9, 12)
sfs.k_score_
0.9791666666666666
pd.DataFrame.from_dict(sfs.get_metric_dict()).T
| feature_idx | cv_scores | avg_score | feature_names | ci_bound | std_dev | std_err | |
|---|---|---|---|---|---|---|---|
| 1 | (6,) | [0.7222222222222222, 0.8333333333333334, 0.742... | 0.76746 | (flavanoids,) | 0.0670901 | 0.0418533 | 0.024164 |
| 2 | (6, 9) | [0.9444444444444444, 1.0, 0.9714285714285714, ... | 0.971825 | (flavanoids, color_intensity) | 0.031492 | 0.0196459 | 0.0113425 |
| 3 | (4, 6, 9) | [0.9722222222222222, 1.0, 0.9714285714285714, ... | 0.985913 | (magnesium, flavanoids, color_intensity) | 0.0225862 | 0.0140901 | 0.00813492 |
| 4 | (4, 6, 9, 12) | [0.9722222222222222, 0.9722222222222222, 0.971... | 0.978968 | (magnesium, flavanoids, color_intensity, proline) | 0.0194714 | 0.012147 | 0.00701308 |
| 5 | (2, 4, 6, 9, 12) | [0.9444444444444444, 0.9722222222222222, 0.971... | 0.972024 | (ash, magnesium, flavanoids, color_intensity, ... | 0.0314903 | 0.0196449 | 0.011342 |
| 6 | (2, 4, 6, 8, 9, 12) | [0.9722222222222222, 0.9722222222222222, 0.971... | 0.978968 | (ash, magnesium, flavanoids, proanthocyanins, ... | 0.0194714 | 0.012147 | 0.00701308 |
| 7 | (0, 2, 4, 6, 8, 9, 12) | [0.9444444444444444, 0.9722222222222222, 1.0, ... | 0.979167 | (alcohol, ash, magnesium, flavanoids, proantho... | 0.0369201 | 0.0230321 | 0.0132976 |
sfs = SFS(RandomForestClassifier(n_estimators=100, random_state=0, n_jobs = -1),
k_features = (1, 8),
forward= True,
floating = False,
verbose= 2,
scoring= 'accuracy',
cv = 4,
n_jobs= -1
).fit(X_train, y_train)
[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 5 out of 13 | elapsed: 2.4s remaining: 4.0s [Parallel(n_jobs=-1)]: Done 13 out of 13 | elapsed: 4.8s finished [2020-08-06 12:57:09] Features: 1/8 -- score: 0.7674603174603174[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 4 out of 12 | elapsed: 2.3s remaining: 4.7s [Parallel(n_jobs=-1)]: Done 12 out of 12 | elapsed: 4.4s finished [2020-08-06 12:57:13] Features: 2/8 -- score: 0.9718253968253968[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 2 out of 11 | elapsed: 1.9s remaining: 8.7s [Parallel(n_jobs=-1)]: Done 8 out of 11 | elapsed: 2.3s remaining: 0.8s [Parallel(n_jobs=-1)]: Done 11 out of 11 | elapsed: 4.3s finished [2020-08-06 12:57:17] Features: 3/8 -- score: 0.9859126984126985[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 7 out of 10 | elapsed: 2.6s remaining: 1.0s [Parallel(n_jobs=-1)]: Done 10 out of 10 | elapsed: 4.4s finished [2020-08-06 12:57:22] Features: 4/8 -- score: 0.9789682539682539[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 4 out of 9 | elapsed: 2.5s remaining: 3.1s [Parallel(n_jobs=-1)]: Done 9 out of 9 | elapsed: 4.3s remaining: 0.0s [Parallel(n_jobs=-1)]: Done 9 out of 9 | elapsed: 4.3s finished [2020-08-06 12:57:26] Features: 5/8 -- score: 0.9720238095238095[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 3 out of 8 | elapsed: 2.1s remaining: 3.5s [Parallel(n_jobs=-1)]: Done 8 out of 8 | elapsed: 2.5s remaining: 0.0s [Parallel(n_jobs=-1)]: Done 8 out of 8 | elapsed: 2.5s finished [2020-08-06 12:57:29] Features: 6/8 -- score: 0.9789682539682539[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 4 out of 7 | elapsed: 1.7s remaining: 1.3s [Parallel(n_jobs=-1)]: Done 7 out of 7 | elapsed: 1.8s finished [2020-08-06 12:57:31] Features: 7/8 -- score: 0.9791666666666666[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 3 out of 6 | elapsed: 1.9s remaining: 1.9s [Parallel(n_jobs=-1)]: Done 6 out of 6 | elapsed: 1.9s finished [2020-08-06 12:57:33] Features: 8/8 -- score: 0.9791666666666666
Let's go ahead and see the accuracy with this 7 features.
sfs.k_score_
0.9859126984126985
Now, we can see here selected feature from this algorithm.
sfs.k_feature_names_
('magnesium', 'flavanoids', 'color_intensity')
Step Backward Selection (SBS)
Let's go ahead work with the Step Backward Selection. Have a look at the following script.
The only thing change here compared to Step Forward Selection, keep forward as False.
sfs = SFS(RandomForestClassifier(n_estimators=100, random_state=0, n_jobs = -1),
k_features = (1, 8),
forward= False,
floating = False,
verbose= 2,
scoring= 'accuracy',
cv = 4,
n_jobs= -1
).fit(X_train, y_train)
[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 5 out of 13 | elapsed: 2.2s remaining: 3.6s [Parallel(n_jobs=-1)]: Done 13 out of 13 | elapsed: 4.6s finished [2020-08-06 12:57:54] Features: 12/1 -- score: 0.9861111111111112[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 4 out of 12 | elapsed: 2.2s remaining: 4.5s [Parallel(n_jobs=-1)]: Done 12 out of 12 | elapsed: 4.5s finished [2020-08-06 12:57:58] Features: 11/1 -- score: 0.9861111111111112[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 2 out of 11 | elapsed: 2.2s remaining: 10.3s [Parallel(n_jobs=-1)]: Done 8 out of 11 | elapsed: 2.7s remaining: 0.9s [Parallel(n_jobs=-1)]: Done 11 out of 11 | elapsed: 4.1s finished [2020-08-06 12:58:03] Features: 10/1 -- score: 0.9791666666666666[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 7 out of 10 | elapsed: 3.1s remaining: 1.3s [Parallel(n_jobs=-1)]: Done 10 out of 10 | elapsed: 4.9s finished [2020-08-06 12:58:08] Features: 9/1 -- score: 0.9861111111111112[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 4 out of 9 | elapsed: 2.1s remaining: 2.7s [Parallel(n_jobs=-1)]: Done 9 out of 9 | elapsed: 4.1s remaining: 0.0s [Parallel(n_jobs=-1)]: Done 9 out of 9 | elapsed: 4.1s finished [2020-08-06 12:58:12] Features: 8/1 -- score: 0.9859126984126985[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 3 out of 8 | elapsed: 2.2s remaining: 3.8s [Parallel(n_jobs=-1)]: Done 8 out of 8 | elapsed: 2.7s remaining: 0.0s [Parallel(n_jobs=-1)]: Done 8 out of 8 | elapsed: 2.7s finished [2020-08-06 12:58:15] Features: 7/1 -- score: 0.978968253968254[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 4 out of 7 | elapsed: 2.1s remaining: 1.6s [Parallel(n_jobs=-1)]: Done 7 out of 7 | elapsed: 2.4s finished [2020-08-06 12:58:17] Features: 6/1 -- score: 0.9859126984126985[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 3 out of 6 | elapsed: 1.7s remaining: 1.7s [Parallel(n_jobs=-1)]: Done 6 out of 6 | elapsed: 1.8s finished [2020-08-06 12:58:19] Features: 5/1 -- score: 0.9789682539682539[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 2 out of 5 | elapsed: 2.1s remaining: 3.2s [Parallel(n_jobs=-1)]: Done 5 out of 5 | elapsed: 2.1s remaining: 0.0s [Parallel(n_jobs=-1)]: Done 5 out of 5 | elapsed: 2.1s finished [2020-08-06 12:58:21] Features: 4/1 -- score: 0.9718253968253968[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 4 out of 4 | elapsed: 2.3s remaining: 0.0s [Parallel(n_jobs=-1)]: Done 4 out of 4 | elapsed: 2.3s finished [2020-08-06 12:58:24] Features: 3/1 -- score: 0.9718253968253968[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 3 out of 3 | elapsed: 2.3s finished [2020-08-06 12:58:26] Features: 2/1 -- score: 0.9718253968253968[Parallel(n_jobs=-1)]: Using backend LokyBackend with 8 concurrent workers. [Parallel(n_jobs=-1)]: Done 2 out of 2 | elapsed: 2.2s finished [2020-08-06 12:58:28] Features: 1/1 -- score: 0.7674603174603174
sbs = sfs sbs.k_score_
0.9859126984126985
Let's get the selected features.
sbs.k_feature_names_
('alcohol',
'malic_acid',
'ash',
'alcalinity_of_ash',
'magnesium',
'flavanoids',
'nonflavanoid_phenols',
'color_intensity')
Exhaustive Feature Selection (EFS)
Let's go ahead and learn about the Exhaustive Feature Selection(EFS).
from mlxtend.feature_selection import ExhaustiveFeatureSelector as EFS
It will start with the subset of minimum features to maximum subset of features.
efs = EFS(RandomForestClassifier(n_estimators=100, random_state=0, n_jobs=-1),
min_features= 4,
max_features= 5,
scoring='accuracy',
cv = None,
n_jobs=-1
).fit(X_train, y_train)
Features: 2002/2002
So, while training with exauhstive feature selection with minimum subset of 4 and 5 it has trained for 2002 subsets.
C(13, 4) + C(13, 5) = 715 + 1287
715 + 1287
2002
Let's find out best accuracy for EFS algorithm with the following code.
efs.best_score_
1.0
Now get the selected features for the best score.
efs.best_feature_names_
('alcohol', 'malic_acid', 'ash', 'alcalinity_of_ash')
Let's get indices of selected features.
efs.best_idx_
(0, 1, 2, 3)
from mlxtend.plotting import plot_sequential_feature_selection as plot_sfs
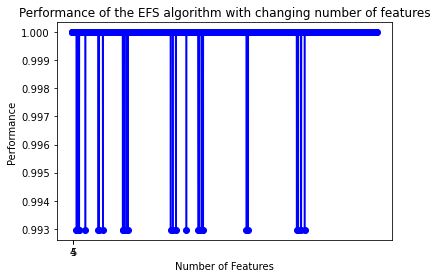
Now, try to plot the graph of the performance with changing number of features.
plot_sfs(efs.get_metric_dict(), kind='std_dev')
plt.title('Performance of the EFS algorithm with changing number of features')
plt.show()
1 Comment